Research
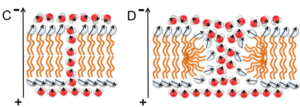
The Computational Biology Group focuses on the structure, dynamics, and function of biological membrane mimics, including e.g. lipid-protein interactions, membrane (nano-)domain formation, and (membrane) protein oligomerization studied both in atomistic and coarse-grained molecular dynamics simulations (Biological Membrane Biophysics).
Current projects include:
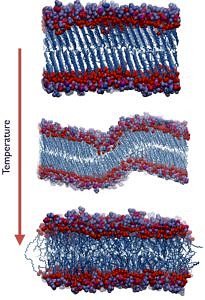 |
Membrane Phase TransitionsWith increasing temperature, lipid bilayers undergo a gel-fluid phase transition, which plays an essential role in many physiological phenomena. Remarkably, the phase transition temperatures of biological membranes were reported to be slightly below the respective physiological temperatures. In this project we address the
L. Sun and R.A. Böckmann. Membrane Phase Transition during Heating and Cooling: Molecular Insight into Reversible Melting. Europ. Biophys. J. 47(2):151-164 (2018) [link] [pdf] |
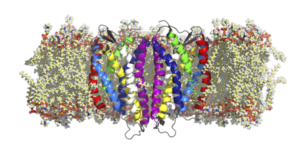
S. Gahbauer, K. Pluhackova, R.A. Böckmann. Closely related, yet unique: Distinct homo- and heterodimerization patterns of G protein coupled chemokine receptors and their fine-tuning by cholesterol.PLoS Comp. Biol. 14(3): e1006062 (2018) [link] K. Pluhackova, S. Gahbauer, F. Kranz, T.A. Wassenaar, and R.A. Böckmann. Dynamic Cholesterol-conditioned Dimerization of the G Protein Coupled Chemokine Receptor Type 4. PLoS Comp. Biol. 12(11):e1005169 (2016) [link] [pdf] |
GPCR Dimerization and Oligomerization.G protein coupled receptors (GPCRs) allow for the transmission of signals across biological membranes. For a number of GPCRs, this signaling was shown to be coupled to prior dimerization of the receptor. In this project, we study the influence of the lipid environment as well as of bound ligands on receptor dimerization and receptor signaling. |
|
Sonja A. Kirsch and Rainer A. Böckmann. Membrane Pore Formation in Atomistic and Coarse-Grained Simulations (Review). BBA-Biomembranes 1858:2266-2277 (2016) [pdf] [link] Rainer A. Böckmann, Bert L. de Groot, Sergej Kakorin, Eberhard Neumann, Helmut Grubmüller. Kinetics, Statistics, and Energetics of Lipid Membrane Electroporation Studied by Molecular Dynamics Simulations. Biophys. J. 95:1837-1850 (2008) [pdf]
|
Membrane Pore Formation.For certain physiological functions e.g. in the fusion of membranes and also in a number of biotechnological applications like gene transfection the membrane integrity needs to be compromised to allow for instance for the exchange of polar molecules across the membrane barrier. Mechanisms enabling the transport of molecules across the membrane involve membrane proteins that form specific pores or act as transporters, but also so-called lipid pores induced by external fields, stress, or peptides. |
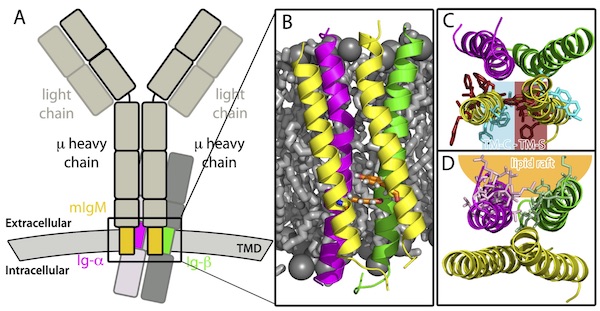
M. Friess, K. Pluhackova, R.A. Böckmann. Structural Model of the mIgM B-cell Receptor Transmembrane Domain from Self-Association Molecular Dynamics Simulations. Front. Immunol. 9:2947 (2018) [link] [pdf]
|
B Cell ReceptorAntigen binding to B-cell antigen receptors (BCRs) followed by signaling initiates the humoral immune response. The signaling is intimately coupled to nanoclustering of BCRs and their sorting to specific membrane domains, a process that is ruled by interactions between the BCR transmembrane domain and lipids. Here, we modeled the structure of the transmembrane (TM) domain of the IgM B-cell receptor using self-assembly coarse-grained molecular dynamics simulations. |